Your Rna primers are removed by the action of the enzyme images are ready in this website. Rna primers are removed by the action of the enzyme are a topic that is being searched for and liked by netizens today. You can Download the Rna primers are removed by the action of the enzyme files here. Find and Download all free vectors.
If you’re searching for rna primers are removed by the action of the enzyme pictures information related to the rna primers are removed by the action of the enzyme interest, you have come to the right site. Our site always gives you suggestions for refferencing the highest quality video and image content, please kindly search and locate more enlightening video content and images that match your interests.
Rna Primers Are Removed By The Action Of The Enzyme. Another enzyme DNA ligase seals the nicks by forming the phosphodiester bond thus generating a continuous sugar-phosphate backbone for the lagging strand. The synthesis of RNA primers is thus guided by the same templating principle used for DNA synthesis see Figures 1-5 and 5-2. RNase H performs three types of cleaving actions. This is the aspect of E.
 Steps And Proteins Involved In Dna Replication Dna Polymerase Mitochondrial Dna Dna Ligase From pinterest.com
Steps And Proteins Involved In Dna Replication Dna Polymerase Mitochondrial Dna Dna Ligase From pinterest.com
More enzymes are needed to clean up the DNA. RNA primers can be removed by DNA polymerase with proofreading activity and RNAseH. After replication the RNA primers are removed by enzyme and replaced with DNA nucleotides. The enzyme that unwinds a segment of the DNA molecule is. The enzyme seals the nicks in the sugar-phosphate backbone after the RNA primers are removed. Termination After the synthesis and extension of both the continuous and discontinued stands an enzyme knows as exonuclease removes all RNA primers from the original strands.
RNase H performs three types of cleaving actions.
The synthesis of RNA primers is thus guided by the same templating principle used for DNA synthesis see Figures 1-5 and 5-2. Once the RNA primer has been removed and replaced the adjacent Okazaki fragments must be linked together. The 3-OH end of one fragment is adjacent to the 5-Phosphate end of the previous fragment. To form a continuous lagging strand of DNA the RNA primer must be eventually removed from the Okazaki fragments and replaced with DNA. In eukaryotic cells RNA primers are removed by the combined action of 5 to 3 exonucleases and a. The removal of RNA primers by DNA polymerase I is an essential part of DNA replication because the final product must consist entirely of double-stranded DNA.
 Source: in.pinterest.com
Source: in.pinterest.com
Coli RNA primers are removed by the combined action of RNase H an enzyme that degrades the RNA strand of RNA-DNA hybrids and DNA polymerase I. The RNA nucleotides from the short RNA primers must be removed and replaced by DNA nucleotides which are then joined by the DNA ligase enzyme. In eukaryotes RNA primers are primarily removed by a. Once the RNA primer has been removed and replaced the adjacent Okazaki fragments must be linked together. Their digestion can only happen in 5 to 3 direction.
 Source: sciencedirect.com
Source: sciencedirect.com
Coli RNA primers are removed by the combined action of RNase H an enzyme that degrades the RNA strand of RNA-DNA hybrids and polymerase I. RNA primers in DNA replication. In eukaryotes RNA primers are primarily removed by a. For DNA polymerase to act it should be extending an existing template and RNA primers should be in their way. Termination After the synthesis and extension of both the continuous and discontinued stands an enzyme knows as exonuclease removes all RNA primers from the original strands.
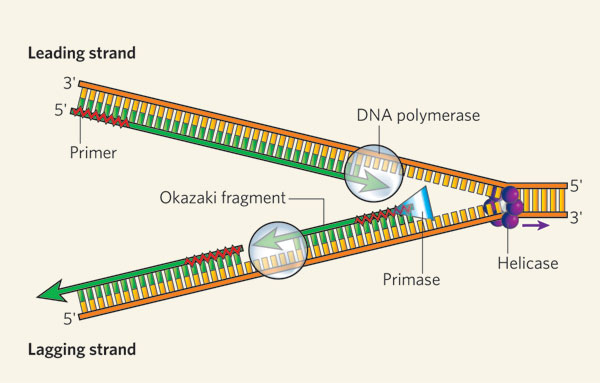 Source: nature.com
Source: nature.com
As synthesis proceeds an enzyme removes the RNA primer which is then replaced with DNA nucleotides and the gaps between fragments are sealed by an enzyme called DNA ligase. Here we show that the endonucleases Dna2 and Fen1 act sequentially to facilitate the complete removal of the primer RNA. RNase H performs three types of cleaving actions. RNA primers can be removed by DNA polymerase with proofreading activity and RNAseH. Because an RNA primer contains a properly base-paired nucleotide with a 3-OH group at one end it can be elongated by the DNA polymerase at this end to begin an Okazaki fragment.
 Source: pinterest.com
Source: pinterest.com
Both work in different conditions. Coli DNA replication in which polymerase I plays a critical role. The last step in the synthesis of the lagging strand of DNA is the formation of a phosphodiester linkage between the 3-hydroxyl group at the end of one Okazaki fragment and the 5-phosphate group of an adjacent Okazaki fragment. RNase H performs three types of cleaving actions. This happens during lagging strand synthesis of DNA replication.
 Source: fi.pinterest.com
Source: fi.pinterest.com
This is the aspect of E. Three major DNA polymerases are then involved. DNA polymerase ɛ loads onto the leading strand and DNA polymerase δ loads onto the lagging strand to synthesize new DNA from the 3 end of iDNA. The synthesis of RNA primers is thus guided by the same templating principle used for DNA synthesis see Figures 1-5 and 5-2. This happens during lagging strand synthesis of DNA replication.
 Source: pinterest.com
Source: pinterest.com
On the lagging strand sections of RNA primer followed by iDNA are removed by an exonuclease and then replaced with new DNA by DNA polymerase δ. DNA polymerase I b. The enzyme seals the nicks in the sugar-phosphate backbone after the RNA primers are removed. Another enzyme DNA ligase seals the nicks by forming the phosphodiester bond thus generating a continuous sugar-phosphate backbone for the lagging strand. In eukaryotes RNA primers are primarily removed by a.
 Source: pinterest.com
Source: pinterest.com
The synthesis of each Okazaki. This is the aspect of E. TF Without the replication of telomeres by the enzyme telomerase linear chromosomes would gradually get longer with each round of replication F TF During replication of a linear DNA molecule the lagging strand can be completely replicated but when the last RNA primer is removed from the leading strand DNA polymerases cannot replace it with DNA. DNA polymerase I d. This is the aspect of E.
 Source: sciencedirect.com
Source: sciencedirect.com
This is the aspect of E. Biology questions and answers. RNA primers can be removed by DNA polymerase with proofreading activity and RNAseH. The Okazaki fragments each require a primer made of RNA to start the synthesis. On the lagging strand sections of RNA primer followed by iDNA are removed by an exonuclease and then replaced with new DNA by DNA polymerase δ.
 Source: pinterest.com
Source: pinterest.com
Ligases are present in both prokaryotes and eukaryotes. The synthesis of each Okazaki. RNA primers in DNA replication. EukaryotesIn eukaryotes a special exonuclease called RNase H appears to work along with other exonucleases to remove the RNA primer in the 5 to 3 directions. The RNA nucleotides from the short RNA primers must be removed and replaced by DNA nucleotides which are then joined by the DNA ligase enzyme.
 Source: pinterest.com
Source: pinterest.com
The strand with the Okazaki fragments is known as the lagging strand. Once the RNA primer has been removed and replaced the adjacent Okazaki fragments must be linked together. Ligases are present in both prokaryotes and eukaryotes. This happens during lagging strand synthesis of DNA replication. As synthesis proceeds an enzyme removes the RNA primer which is then replaced with DNA nucleotides and the gaps between fragments are sealed by an enzyme called DNA ligase.
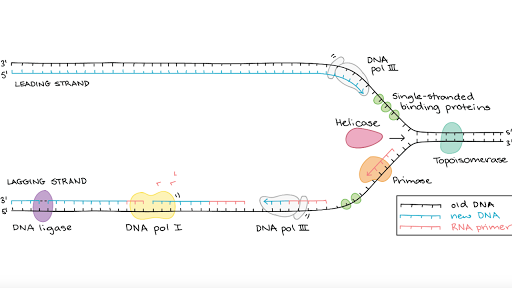
The responsible for sealing this nick lies with the enzyme DNA ligase. TF Without the replication of telomeres by the enzyme telomerase linear chromosomes would gradually get longer with each round of replication F TF During replication of a linear DNA molecule the lagging strand can be completely replicated but when the last RNA primer is removed from the leading strand DNA polymerases cannot replace it with DNA. More enzymes are needed to clean up the DNA. Both work in different conditions. RNA primers in DNA replication.
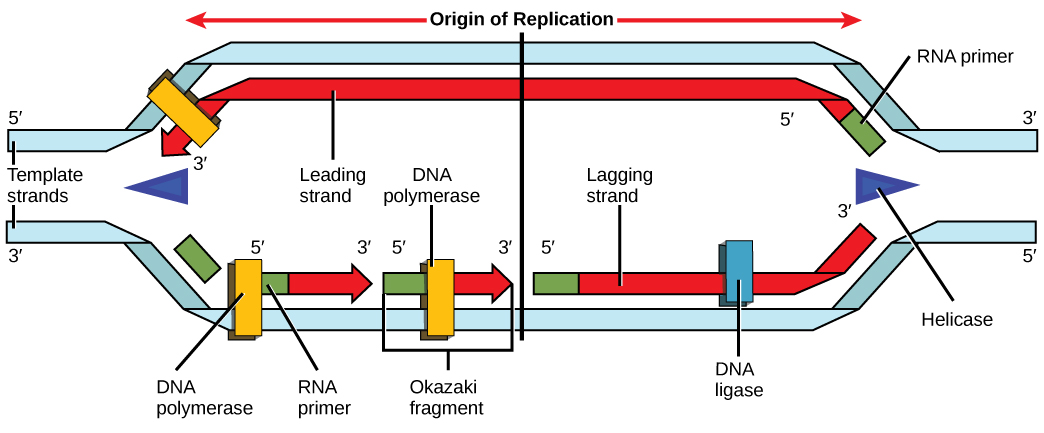 Source: opentextbc.ca
Source: opentextbc.ca
α δ and ε. DNA Polymerase III synthesizes the majority of the DNA while DNA Polymerase I synthesizes DNA in the regions where the RNA primers were laid. This is the aspect of E. DNA polymerase α e. Termination After the synthesis and extension of both the continuous and discontinued stands an enzyme knows as exonuclease removes all RNA primers from the original strands.
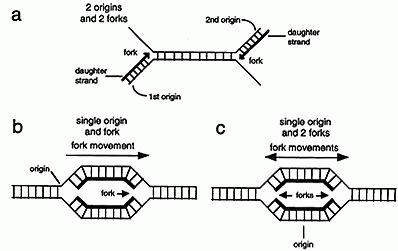 Source: www2.csudh.edu
Source: www2.csudh.edu
Non-specific degradation of the plus-strand RNA genome specific removal of the minus-strand tRNA primer and removal of the plus-strand purine-rich polypurine tract PPT primer. α δ and ε. Another enzyme DNA ligase seals the nicks by forming the phosphodiester bond thus generating a continuous sugar-phosphate backbone for the lagging strand. The synthesis of each Okazaki. More enzymes are needed to clean up the DNA.
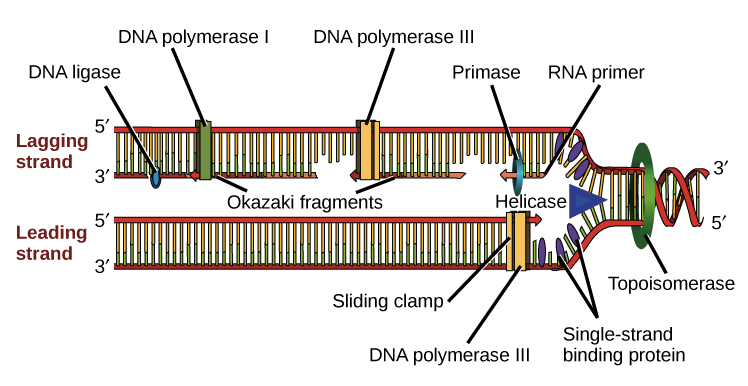 Source: khanacademy.org
Source: khanacademy.org
DNA polymerase I removes the RNA primer and fills in the gaps with DNA. To form a continuous lagging strand of DNA the RNA primer must be eventually removed from the Okazaki fragments and replaced with DNA. DNA polymerase I d. EukaryotesIn eukaryotes a special exonuclease called RNase H appears to work along with other exonucleases to remove the RNA primer in the 5 to 3 directions. After replication the RNA primers are removed by enzyme and replaced with DNA nucleotides.
 Source: pinterest.com
Source: pinterest.com
In eukaryotes RNA primers are primarily removed by a. This is the aspect of E. RNA primers are removed by the action of the enzyme. The synthesis of each Okazaki. Coli RNA primers are removed by the combined action of RNase H an enzyme that degrades the RNA strand of RNA-DNA hybrids and polymerase I.
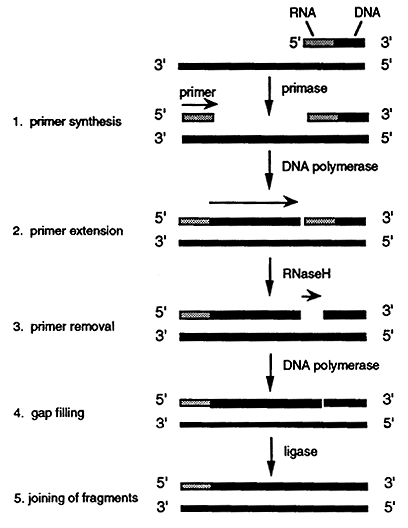 Source: www2.csudh.edu
Source: www2.csudh.edu
The removal of RNA primers by DNA polymerase I is an essential part of DNA replication because the final product must consist entirely of double-stranded DNA. RNA primers are removed by the action of the enzyme. Another enzyme DNA ligase seals the nicks by forming the phosphodiester bond thus generating a continuous sugar-phosphate backbone for the lagging strand. This is the aspect of E. Coli DNA replication in which polymerase I plays a critical role.
 Source: nature.com
Source: nature.com
For DNA polymerase to act it should be extending an existing template and RNA primers should be in their way. Coli RNA primers are removed by the combined action of RNase H an enzyme that degrades the RNA strand of RNA-DNA hybrids and DNA polymerase I. The sequential action of. DNA Polymerase III synthesizes the majority of the DNA while DNA Polymerase I synthesizes DNA in the regions where the RNA primers were laid. The removal of RNA primers by DNA polymerase I is an essential part of DNA replication because the final product must consist entirely of double-stranded DNA.
 Source: pinterest.com
Source: pinterest.com
On the lagging strand sections of RNA primer followed by iDNA are removed by an exonuclease and then replaced with new DNA by DNA polymerase δ. To form a continuous lagging strand of DNA the RNA primer must be eventually removed from the Okazaki fragments and replaced with DNA. The removal of RNA primers by DNA polymerase I is an essential part of DNA replication because the final product must consist entirely of double-stranded DNA. Termination After the synthesis and extension of both the continuous and discontinued stands an enzyme knows as exonuclease removes all RNA primers from the original strands. DNA polymeraseδ then returns to synthesize DNA where the RNA primer was.
This site is an open community for users to share their favorite wallpapers on the internet, all images or pictures in this website are for personal wallpaper use only, it is stricly prohibited to use this wallpaper for commercial purposes, if you are the author and find this image is shared without your permission, please kindly raise a DMCA report to Us.
If you find this site adventageous, please support us by sharing this posts to your favorite social media accounts like Facebook, Instagram and so on or you can also save this blog page with the title rna primers are removed by the action of the enzyme by using Ctrl + D for devices a laptop with a Windows operating system or Command + D for laptops with an Apple operating system. If you use a smartphone, you can also use the drawer menu of the browser you are using. Whether it’s a Windows, Mac, iOS or Android operating system, you will still be able to bookmark this website.






